Standard PCR protocol , how to do PCR (Polymerase Chain Reaction) 15th November 2021 – Tags: PCR, PCR protocol, Polymerase chain reaction
Introduction
This article will discuss the PCR protocol in depth to provide clarity on this topic.
Once you have your genetic samples, one of the first steps in PCR is to design the primers needed to perform the PCR reaction.
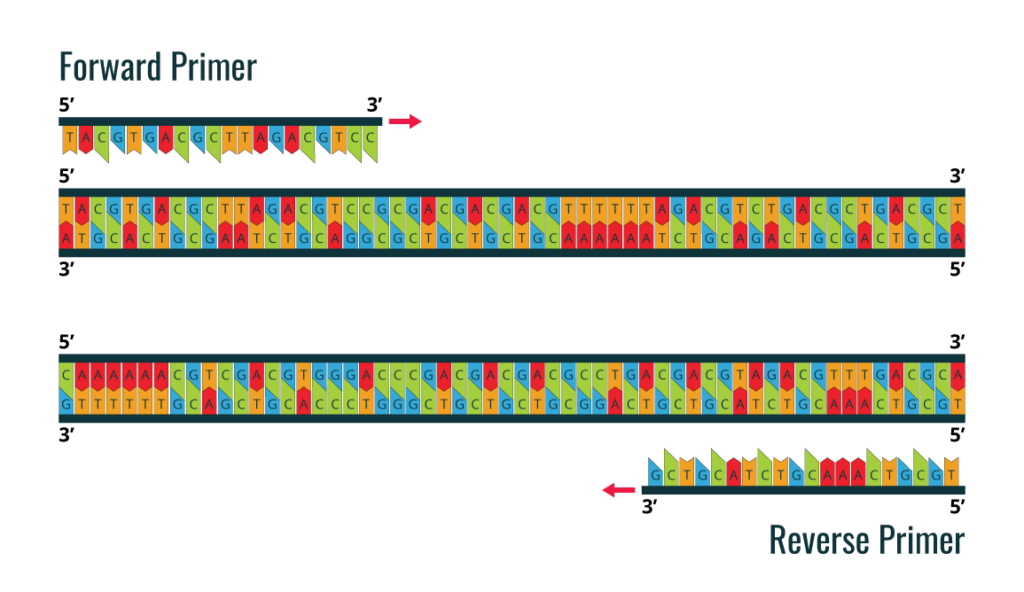
The first step in proceeding with a standard PCR is to design the primers. You will need to determine which fragment of the DNA template you want to amplify, for which you will need to know the DNA sequence of the template. The NCBI database, a web server with all known DNA sequences, is a good resource you can use to search for your sequence. Next, you will need to design two primers, the forward and reverse primer. Primer design is a critical step in a PCR protocol. The primer set must flank the fragment you intend to amplify from the DNA template. The direct primer will bind to the 3′-5′ DNA strand and the reverse primer will bind to the 5′-3′ DNA strand. See the diagram below.
Cebador delantero o cebador inverso
Many PCR protocol or PCR protocol problems are associated with incorrect primer design. The most common design errors include
- Primer dimers
- Primer self-annealing hairpin loop structures
- Non-specific annealing
The length of the primer should be about 18-30 bp, the GC content close to 50% and the melting temperature (TM) between 55°C and 65°C. To calculate the melting temperature, the following equation Tm = 2°C(A+T) + 4°C(G+C) can be used. Although most commercial DNA polymerases provide their own calculator and specific instructions for different conditions, there are several web-based platforms to help you design primers.
Once you have obtained your DNA samples and designed your forward and reverse primers, you can proceed with the PCR experiment.
General PCR Protocol
Reagents
- Direct and reverse primers
- dNTPs
- DNA Template
- DNA polymerase
Note: From the beginning of your PCR experiment to the end, you should always wear gloves to avoid DNA contamination. All reagents, primers and enzymes must be kept on ice. Make sure that the primers, DNA template and buffer are completely thawed before you start preparing the PCR solution.
It is important to create an experimental design according to scientific guidelines, including a positive control and a negative control. As a negative control, you can prepare a private PCR DNA template. For the positive control, you should use a set of primers and DNA template that has been shown to work well in previous experiments.
Depending on the DNA polymerase you use, the final concentration of each reagent will vary. These are the standard volumes and concentrations for a 50ul reaction:
- In 200 μl PCR tubes:
- Add 38 μl of sterile water.
- Add 2 μl of direct primer (10 μM).
- Add 2 μl of reverse primer (10 μM).
- Add 1 μl of dNTPs (50 μM).
- Add 5 μl of reaction buffer with MgCl2 (10X).
- Add 1 μl of DNA template (100 ng/μl).
- Add 1 μl of DNA polymerase (0.5 U/μl).
Note: If you are willing to perform a large number of PCR reactions, it is recommended to prepare a reaction mixture, excluding reagents that will be different from one experiment to another (typically the DNA template or primer set).
- Gently pipette the reaction mixture to allow good homogenisation.
- A short spin in a centrifuge is recommended.
- Make sure that the caps of the PCR tubes are closed.
- Insert the tube into the thermal cycler.

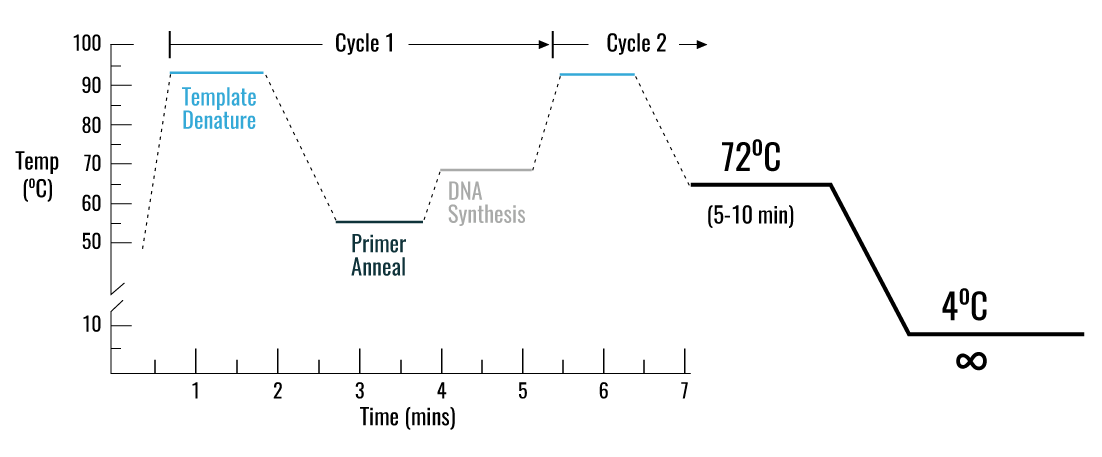
The PCR protocol to follow is:
Nota: Always pay attention to the guidelines of the DNA polymerase, as the temperature and time of denaturation, annealing and extension are highly dependent on it. On the thermal cycler, you will need to set these steps later (see diagram above):
- An initial DNA denaturation step at 94°C to 98°C for 3 to 5 minutes, depending on the optimum temperature of the DNA polymerase.
- Denaturation temperature (same as used in the previous step) for 30 seconds
- Annealing temperature taking into account the Tm of the primers for 30 seconds
- Extension temperature at 72°C and taking into account the size of the fragment to be amplified.
These steps must be repeated for 25 to 35 rounds (cycles). A final extension step is required to allow all PCR products to be synthesised correctly, usually at 72°C for 10 min. Finally, the temperature should be reduced to 4°C to store the PCR product.
PCR analysis: Gel electrophoresis
- Load the 6 μl mixture onto a 1% agarose gel.
- Load 5 μl of DNA marker on the same gel.
- Use a UV transilluminator to visualise the PCR product on the agarose gel.
Note: To avoid ethidium bromide staining, you can use pre-stained Midori or Red Safe gels, which are less toxic compounds. Use a DNA marker to compare the correct size of the PCR product.
Reverse transcription protocol
Reverse transcription is the process by which RNA is transcribed into DNA, which will allow us to perform experiments such as qRT-PCR under the catalytic activity of a specific DNA polymerase that is only capable of amplifying fragments of DNA molecules.
Note: From the beginning of your PCR experiment to the end, you should always wear gloves to avoid DNA contamination. All reagents, primers and enzymes should be kept on ice.
- In a 200 μl PCR tube:
a. Add 2 μg of total RNA.
b. Add 1 ul of oligo dt (50μM).
c. Add 1 μl of dNTP (10mM).
d. Add DEPC-treated water up to 10 μl. - Incubate at 65 °C for 5 min and then keep the tubes on ice for 1 min.
- Add 2 ul of RT buffer (10X).
- Add 4 μl of MgCl2.
- Add 2 μl of DTT (0.1 M).
- Add 1 ul of RNase inhibitor (40 U/μl).
- Add 1 ul of reverse transcriptase (200 U/μl).
- Incubate at 50°C for 60 min, followed by 85°C for 5 min.
- Afterwards, keep the tube on ice.
- Add 1 μl of RNase H.
- Incubate for 30 min at 37°C.
- Store the PCR tube at -20°C.
- For RNA quantification and quality, use a NanoDrop kit to measure RNA (260 nm), protein and salt concentrations.
qRT-PCR PROTOCOL
Similar to standard PCR, a set of primers as described above should be designed to amplify each cDNA fragment of interest, which usually corresponds to a specific gene. In addition, it is also recommended to design specific primers for other cDNA genes for data normalisation procedures, which are also known as “housekeeping” genes (e.g. 18S, GAPDH, ACTB).
For each cDNA gene, prepare the following reaction mixture in 200 μl PCR tubes for a final volume of 50 μl:
- Add 25 μl of SYBR Green Mix (2x).
- Add 1 μl of cDNA from the previous cDNA sample.
- Add 1 μl of Forward Primer (10 mM).
- Add 1 μl of Reverse Primer (10 mM).
- Add 22.5 μl of DEPC water.
- Insert the tubes into the thermal cycler.
After completion the PCR Protocol, perform quantification analysis by the comparative Ct method. The Ct method compares the Ct values of the samples with the standard samples. The Ct values of controls and samples are normalised using the Ct values obtained for the housekeeping genes. To validate the Ct calculation, the target amplification efficiencies are required to be close to the endogenous ones. This can be achieved by using different dilutions of the template.
Mouse IFN-beta ELISA
Features Mouse IFN-beta ELISA
Optimised capture and detection antibody pairings with recommended concentrations save significant development time
Development protocols are provided to guide assay optimisation
Assay can be tailored to your specific needs
Cost-effective alternative to complete kits
SOCS-1 Antibody
SOCS1 is a member of the STAT inhibitor (SSI) family, also known as suppressors of cytokine signalling (SOCS). Members of the SSI family are negative regulators of cytokine signalling. SOCS1 functions downstream of cytokine receptors and participates in a negative feedback loop to attenuate cytokine signalling.
Polyclonal antibodies are produced by immunising animals with a synthetic peptide corresponding to the residues surrounding Ala156 of human SOCS1. The antibodies were purified by protein A and peptide affinity chromatography.
Anti-SLC1A5 Antibody
Several suppliers offer anti-SLC1A5 antibodies. This gene encodes the solute carrier family 1 member 5 protein in humans and may also be known as ASCT2, AAAT, ATBO, M7V1, neutral amino acid transporter B(0), and ATB(0). Structurally, the protein has a mass of 56.6 kilodaltons. Based on the gene name, canine, porcine, monkey, mouse and rat orthologues can also be found.
References
- “Sinobiological Rabbit Polyclonal Antibodies – Rabbit pab”
- What are Polyclonal Antibodies (pAbs)? By Susha Cheriyedath, M.Sc. Reviewed by Deepthi Sathyajith, M.Pharm.
- “Customized polyclonal antibodies“
- AN OVERVIEW OF POLYCLONAL ANTIBODIES AND THEIR APPLICATIONS
- Overlooked benefits of using polyclonal antibodies
Read much more on our blog